|
NOTE: LabAnalyst will open double-clicked or ‘dragged and dropped’ data files of appropriate formats and extensions (.whog, .whsc, .WHtext, .WHdata, .WHSable). 'Raw' Sable-format (ExpeData) files imported from Windows systems have the extension .exp and are not recognized by LabAnalyst. However, if you change that to .expS, these files show with a new icon (see below) and can be opened by drag-and-drop or double-clicking. 'Raw' .exp files can be opened with the Import... option of the FILE menu (see below)
The program can have up to 20 data files open simultaneously, each in its own window. You can switch between files either by clicking the appropriate window, or with the WINDOW menu. Individual files can be closed either by clicking the standard 'close' button, or if the window of the file you want to close is active (in front), with the close file option in the FILE menu.
If you have the maximum number files open and attempt to load another, you will be asked which of the currently open files you want to close to make room for the new file.
For any file format, you can automatically scan and fix large ‘spikes’ in the data by selecting the ‘Automatically Filter Newly-loaded Files’ option in the PREFERENCES/FileHandling menu.
OPEN WARTHOG...  O Loads data files produced by LabHelper
or LabAnalyst. There are four types: O Loads data files produced by LabHelper
or LabAnalyst. There are four types:
|

| Warthog-format
text files (old and slow; retained
for legacy data) |
|
|

| raw binary chart data
extension .whog |
|
|

| raw binary scope data
extension .whsc |
|
|

| binary edited data (saved from LabAnalyst)
extension .WHdata |
|
The file opening window highlights only these types with the proper extensions. Three are binary formats, for LabHelper
and edited LabAnalyst files: the older
Binary-Coded Decimal (BCD) and Floating-Point (FP). LabAnalyst
will transparently read either, but will write only in
FP format. The main difference is speed: BCD
files load more slowly (since they are read and converted one sample
at a time), while FP files load quickly (since entire channels
are read directly from disk into memory).
If you click the toolbar 'Open' or 'Append' or 'Stack'
buttons, you will get this window if you've not specified
in the 'Preferences' window that
only Warthog files will be shown:

This gives you the choice of Warthog (current macOS files or 'classic' files obtained on earlier Mac OS versions [OS8, OS9, etc]), ASCII (text) or Sable SSCF or ExpeData files
(see below).
Two of the buttons ('Warthog Only') and ('Sable or Warthog Binary') will only access files with the correct extensions (.whog, .whsc, .WHdata, .exp, .expS, .WHsable). The 'Any Sable or Warthog File' button allows access to all file types, but only files in Warthog or Sable format will load properly. If you attempt to load a file that isn't Warthog or Sable format, you'll get an error message.
IMPORT
 This
submenu has three items: This
submenu has three items:
WARTHOG BINARY FROM 'CLASSIC' OS9... For old
Warthog binary files from prior to macOS. WARNING: only files with these formats will load properly.
 Note: binary files from 'Classic' Macs (OS8, OS9) may look like this:
Note: binary files from 'Classic' Macs (OS8, OS9) may look like this: You can make the correct icons appear by adding the appropriate file extension (e.g., '.whog' ).
|
TEXT [ .csv, ASCII ] FILE... This routine lets you load data in text (.csv) files, as long as:
- data are in spreadsheet format with variables (columns) delimited by commas or tabs (spaces do not work as delimiters) and cases (rows) delimited by carriage returns
- the data are numeric only (text data are ignored)
- the input file may contain many variables but you must select a maximum of 40.
- NOTE: this will NOT import complex Excel worksheets (i.e., .xls; .xlsx). If you want to load data in Excel format into LabAnalyst, save a copy as a tab-delimited text file (.txt) or .csv file, which LabAnalyst can read.
Go to this page for more information on importing text files.
SABLE
FILE... Loads Sable
Systems data files in SSCF format or the similar but newer 'ExpeData' format. Nearly all Sable files will load;
those which have more than 40 variables will load the first 40 only. The maximum number of markers in
LabAnalyst is about 20,000; Sable files can
(theoretically; extremely rarely in practice) exceed that limit. The converter will read the first 20,000
markers only. Older SSCF files do not include channel labels, so after
the completion of file loading you are prompted to provide your own labels.
Also, the values for mass, flow rate, effective volume, and so forth are
arbitrary and may have to be edited, and there is no information on voltage conversions. The newer ExpeData files contain channel labels and many conversion factors.
NOTE: Because of the need to interconvert numeric formats for every data point, large Sable files (e.g. with > 10,000 cases) may be slow to load. Very. Slow. You may see the dreaded 'beachball', but if you ignore it and wait, your patience should eventually be rewarded.
| If you save Sable files from LabAnalyst or LabHelper,
they will appear (on a Mac) with a custom icon (extension .WHSable): |
 |
Provided that LabAnalyst has been run at least once, if you place a raw Sable file on your computer and change the standard .exp suffix to .expS,
it will appear with this icon:
(The file itself is unchanged) |
 |
Back to top
APPEND OR STACK
 This
submenu has 3 items: This
submenu has 3 items:
WARTHOG
FILE...
SABLE FILE... Merges
a Warthog-format file (either text or binary) or a Sable SSCF file
with the currently loaded file. The new file can either be appended to the 'right' (i.e., adding new cases) of the current file, or stacked to the 'bottom' (i.e., adding new channels).
Appending is much more common than adding channels. In either case, the following window opens (appearance will vary depending on the number of channels and samples in the two files):

The program checks to
make sure the merged file will not exceed the current maximum file size -- unlikely unless one or both of the files are very large (a warning
is shown if this is the case). However, the program does not check to make
sure the channel contents match in the current and new files. Thus it is
possible to create a new file that contains two (or more) types of
data in a single channel (i.e., a set of temperature data from the
initial file, then gas concentration data from a second file, then
wind speed data from a third file, etc.).
Needless to say, this can cause considerable confusion unless
you are careful to avoid merging files with differing channel types.
To help avoid such mistakes, the computer provides a graphical display
of the old and new data. In this example, the file to be merged has
20 channels and the current file contains 21 channels.
LabAnalyst will allow access to
the maximum number of channels in either the current or new file.
Sable SSCF files don't contain channel labels, so if you merge them you HAVE
to know ahead of time what the channel structure is. It's safer to first save them into a Warthog-format version.
TIME SCALE ADJUSTMENT... This is a specialized operation that has to be used with care. It is primarily intended to equalize the effective sampling rates of two files that you want to merge or append. This is done in one of two ways, depending on the old and new sampling rates. It is important to note that even if you add points, you do NOT add information. And if you make the file smaller, information will be LOST. Therefore, it is a good idea to make a copy of the file before you perform this manipulation.
• If the new rate is “faster” than the old rate (i.e., more samples per unit time), LabAnalyst will add the extra data points by interpolation. For a simple example, if the original sample rate was one Hz (one sample per second) and you want to change that to two Hz (two samples per second), the program will fit one additional data point between each of two successive existing points. The new point will have a value that is the mean of the two points surrounding it. The operation is similar, but more complex, if the new and old sample rates are not integral multiples of each other.
As mentioned earlier, don’t make the mistake of thinking that because you have added more data points, you somehow have added information content.
• If the new rate is “slower” than the old rate (i.e., fewer samples per unit time), LabAnalyst will simply remove data points as appropriate. It does not do any averaging. Consequently, this operation reduces the amount of information in the file…. so use with care (and make backup copies if you are wise).
These operations are transient unless you save the file (or use the ‘Save as’ option to make a copy with a different name). But if you save a time-adjusted file under its original name, the file is permanently changed. You cannot ‘undo’ time adjustment. The time scale adjustment window looks like this:

Back to top
Screen display basics
After a file is loaded, the 'plot area' in the top part of the screen displays
either the first channel of the file (single
channel mode) or several user-selected channels (multi-channel
mode); see the VIEW menu for details.
The screen in single-channel mode looks approximately as shown below
-- but note that immediately after loading, no block will be selected and
no data will appear in the results or block windows.
The colors of the plot
area and block window are user-selectable (VIEW
menu) and may differ from this example.
 The major components are the plot area (top half of the screen), yellow
data bar (upper left), screen selection slider (bottom edge of plot area), 'toolbar'
menu buttons (across the center, just below the screen selection
slider -- optionally, the toolbar can be above or to the left of the plot area), comments window (middle right), block window (lower
right), and results window (lower left; this contains controls for EDIT menu operations as well as analysis results). A selected block
appears as a color-inverted segment of the plot area. Markers entered during acquisition appear just above the plot area; small 'n' buttons along the top or bottom of the plot area (not in this example) indicate notes entered while gathering data; clicking one of these buttons brings up the note text (example):
The major components are the plot area (top half of the screen), yellow
data bar (upper left), screen selection slider (bottom edge of plot area), 'toolbar'
menu buttons (across the center, just below the screen selection
slider -- optionally, the toolbar can be above or to the left of the plot area), comments window (middle right), block window (lower
right), and results window (lower left; this contains controls for EDIT menu operations as well as analysis results). A selected block
appears as a color-inverted segment of the plot area. Markers entered during acquisition appear just above the plot area; small 'n' buttons along the top or bottom of the plot area (not in this example) indicate notes entered while gathering data; clicking one of these buttons brings up the note text (example):

The appearance of the plot area depends on the screenwidth
(the width of the plot window in pixels) and the number of cases. If the
file contains <= screenwidth cases,
data are plotted 1 pixel per case (if the number of cases is < 50% of
screenwidth, the plot is expanded,
with more than 1 pixel per case). If the total number of cases is
more than screenwidth, the X-axis will
be scaled to fit within the screen dimensions. 'Screens' (consisting
of screenwidth cases) are indicated
by vertical tick marks in the slider.
The example shows a file containing many screenwidths
of data.
If there are more than screenwidth
cases, the bottom of the plot area will show the screen selection slider.
The currently 'active' segment of the file -- the active screen
- is indicated by the slider position. Use the slider or the left
and right arrow keys to change the active screen. To view the active
screen only (necessary for some manipulation and analysis operations), hit
the shift or return keys. Do this again to go back to
viewing the entire file (this example shows 'entire file' mode).
Any markers are shown as vertical dashed lines. While in the plot area, the cursor is a cross-hair; out of
the area it is usually an arrow. The data bar has a readout of case number,
elapsed time, and the value of the currently active channel at the cursor
position. The time of day is shown (24-hour scale) along the bottom of the
plot area, with optional hour indicators (vertical dotted lines).
To perform analyses and some transformations, you need to select a data
block. This can be done by the normal 'click-hold-and-drag'
method (the cursor must be within the plot area). A second method
uses individual clicks: move the cursor to the desired start point and
click once; repeat for the end point (which may be on either side
of the start point). Alternately, blocks of defined width can be selected
with single clicks (see the BLOCK
WIDTH option in the ANALYZE
menu). If there is sufficient space, you can 'shift' blocks to the left or right with the BLOCK
SHIFT commands (also in the ANALYZE
menu).
You can select blocks in 'entire file' or in 'active screen only' mode, and blocks can include multiple screens. If a single
screen is shown, mark the start with a single click, then shift
screens until the end can be marked.
A selected block is indicated as a color-inverted rectangle in the plot
area, and is scaled to fit within the block window (discrete data points
on the block window only appear if the number of included points is small).
Back to top
| Note for file saving options: When you save a file in either Sable or Warthog binary format, with either the Save... or Save As... options, LabAnalyst will add a file extension ( .WHdata for Save...; .WHSable if you select Sable-format output in the Save As... option). The new extension will replace any existing extension, which in effect creates a new file if the source file had a different extension (e.g, .whsc, .whog, .sabl, .ext, .extS) or no extension.
|
SAVE...
⌘S Stores
the current file including any modifications you made. Note
that binary files saved from LabAnalyst
have different icons than the 'raw' data files generated by LabHelper
.
SAVE
AS... Stores the current file -- or optionally
a marked block -- with any modifications you made. You may save all
channels or a subset. Files are stored in Warthog binary (file type 'WHog')
format, in Sable format (ExpeData), or as text files (comma-separated values or tab-separated values). The 'standard format' button saves the existing file (with any modifications
you have made with transformations, smoothing, etc.) under a new file name of your choice.
If 'standard format' was not clicked, after selecting the channels to save, the file format selection window appears, as shown below:
| As mentioned above, Sable ExpeData files do not include values for mass, flow rate, barometric pressure, temperature,
and effective volume (unless written in the comments), and comments are limited to 240 characters. |
| The 'Include Time Index With Each Sample' option saves the real time of each sample in hours (24-hour format). For example, a sample read at 5:45 PM would be indicated as a time of 17.750. Additionally, the elapsed time (in seconds) since the first sample is saved in another column. |
| The 'Time Counter' option creates a variable that indicates elapsed time in hours. For example, this variable would have the value of '1' for all data during the first hour, '2' for all data during the second hour, and so forth. |
| If you are saving in Text format, you can save any markers in the file in a separate column. |
| The 'Append Metadata To End Of Text File' option will save the sample rate, sample number, duration, start time, start date, number of channels, and number of markers after the last line of data. |
| 
|
If you select either of the text file options, you have two formatting choices:
- Timing data are stored with each case (elapsed time since acquisition began, and time of day). This option is useful
for sending files to spreadsheets or plotting programs where time data
are important.
- The sample interval and start time are stored in the comment string
(the first line in the file).
- You can also perform a 'conditional' save, which lets you save only those cases that meet a Boolean selection criterion (see below)
- NOTE: if you use conditional save, the time information (time of day, etc.) in the file will no longer be accurate. Therefore, it is highly recommended that you make backup copies of the original data before the conditional save operation.
The 'Conditional save' option lets you filter data case by case and save only the cases that meet the selection conditions. You need to specify which channel is to be tested (it is not necessary that the tested channel will be saved), one of five Boolean criteria (<, =<, =, >=, >), and a selection criterion (a number against which the individual case values in the tested channel are compared).
In this example, only cases where the value of channel 13 (wheel#1 speed in RPM) is greater than 0.5 will be saved. |
| 
|
Data files (other than text format) that have been saved by LabAnalyst have these icons:

binary files (.WHdata)
|

Sable formats (.WHSable)
|
LabAnalyst saves some other file types, shown here (these may look different -- or may not appear -- on your system, depending on what version of macOS is running):

Preference files
(.LApr)
|

Script files
(.LAscript)
|
EXPORT BLOCK AS csv This is a 'shorthand' version of the SAVE AS option. It saves the selected block of data as a .csv (comma-separated values) text file, readable by Excel and many other programs. You can select which channels to save but otherwise the file format is fixed, with date (Julian date; day of year) and elapsed time included with every data point.
FILE INFORMATION Opens a window
showing the labels of all the channels, the sample interval, the time and
date of storage, etc., and the conversions used by the LabHelper
acquisition program to change raw voltages into appropriate units (temperature,
gas concentrations, speed, etc.). The type of conversion equation
("transform") and the number of samples averaged for each recorded point
("N") are also shown. Except for early data files (circa 2003 or prior), the A-D converter used is also shown (you can add this information if necessary with the popup menu in the lower right, followed by saving the file).
Note that a file loaded in Sable or ASCII format may not show all of the variables.
The following example shows the file data for a 13 channel file (this was recorded prior to implementation of A-D channel storage in data files).

Conversion equations:
Most conversion equations used by the Warthog acquisition program LabHelper
are 2-order polynomials (shown as "poly" in the transform column):
value = A + B*volts + C*volts2
Note that a C of zero produces
a linear conversion. Occasionally a power function is used:
value = A + B*voltsC
Note that in a power function a non-integer C will produce meaningless data if the voltage is negative;
in this condition, LabHelper sets the
results to zero. A C value
of 1 produces a linear conversion.
LabHelper allows use of the keyboard
as an event recorder, and a single channel can use a 3-degree
polynomial conversion. If data have been transformed or copied
into a new channel, no coefficients are shown.
PRINT METADATA Prints a simple text-format listing of the file's metadata (all the sampling criteria, voltage conversions, etc. shown in SHOW FILE DATA (above), along with any user comments). This option can also be accessed in the SHOW FILE DATA window, in the 'Edit Comments' option in the EDIT FILE DATA window (in the EDITmenu), or by clicking the 'Expand' button in the small comments window.
SUMMARY STATS Shows each channel's data range and mean, as in the example below. The summary can be sent to an .xls or .csv format file with the 'Print to Spreadsheet' button.

Back to top
PRINT FILE IMAGE... Saves an image of the
file as a .png file, which you can optionally send to a printer or turn into a .pdf. You can usually do this by hitting the
's' key. If you have selected a block, you have the option of printing
an image of the block or of the entire file.
The .png file
will match whatever is shown in the on-screen viewing mode (Entire
File or Active Screen). A file displayed in the "Compacted
and Averaged" format will be printed in that format. If a
block has been selected, you have the option of printing only the block.
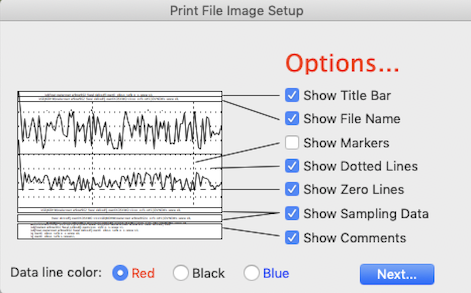
Using the window shown above, you can select which features will appear in the image, such as labeling,
file information, comments, and markers. If the file has more than
one channel, you can print any subset of the channels.
If you click the 'non-standard
plot heights' button, a window will open to allow customization of the
height of each printed channel, as in the image below:

The window contains a notional diagram of the printed page. You select the
relative height of each channel in sequence by moving the cursor on the
diagram to the desired height, and then clicking the mouse ONCE. The
channels will be redrawn one by one as their heights are selected.
Note that there is a fixed amount of room on the page, so that enlarging
one channel requires shrinking one or more of the others. Consequently,
the computer reserves an amount of space necessary for plotting the remaining
channels at the minimum possible height (and will not let you exceed that
limit). You must select plot heights for all the channels. When
done, you can accept the results, redo the plot height selection, or revert
to the normal setting (all channels plotted with equal heights).
Back to top
SEND RESULTS TO
 This
submenu has two items. This
submenu has two items.
SPREADSHEET... Opens
a spreadsheet-format text file for storing results generated
by analysis menu operations. Data are saved in a tab-delineated format
readable by many spreadsheet programs or statistical packages. Optionally,
you can save results to an Excel-format
file (double-clickable directly into Excel).
Besides the results data, the spreadsheet file also contains column labels.
Mean, SD, N, and variable name are always saved; other parameters (start
of block, end of block, start and end time, elapsed time, analysis type,
mass, and channel number and file name) are user-selectable. You can also
enter one or two user-typed 'notes' with each case (edit fields
appear in a window over the COMMENTS window when the 'p' key is struck;
hit 'return' or click the window close button when the values are correct).
Certain analyses, such as regressions, do not fit this format and cannot
be put into spreadsheets.

Storage of results (in memory) occurs when you press the 'p' key (with
the Results window open), or select OUTPUT RESULTS. The spreadsheet
file is not saved to disk until you exit from LabAnalyst
or use the PRINT | CLOSE FILE option.
Some additional considerations:
- Certain analyses, such as regressions and integration, do not fit this
format and cannot be put into tabular files.
- When doing waveforms or pairs-difference, pressing the 'p'
key stores BOTH frequency and amplitude data. If you want to store
one or the other, press the 'f' key to store frequency data only, or the
'a' key to store amplitude data only.
SIMPLE TEXT
FILE... Opens a text file for storing the contents
of the Results Window. The format of the disk file is similar to
that shown in the Results Window and is identical to the format of data
sent to the printer. These files are not formatted for use in spreadsheet-based applications, such as Excel.
Analysis results are transferred to temporary storage in a memory table when you press the 'P' or 'p' key, or select OUTPUT RESULTS.
Real storage (writing data to disk) occurs when you select the
PRINT | CLOSE FILE ( ⌘J ) option. However, any temporary memory tables are automatically saved to disk if you quit the program.
Back to top
|


 O Loads data files produced by LabHelper
or LabAnalyst. There are four types:
O Loads data files produced by LabHelper
or LabAnalyst. There are four types: This
submenu has three items:
This
submenu has three items:
 This
submenu has 3 items:
This
submenu has 3 items:
 The major components are the plot area (top half of the screen), yellow
data bar (upper left), screen selection slider (bottom edge of plot area), 'toolbar'
menu buttons (across the center, just below the screen selection
slider -- optionally, the toolbar can be above or to the left of the plot area), comments window (middle right), block window (lower
right), and results window (lower left; this contains controls for EDIT menu operations as well as analysis results). A selected block
appears as a color-inverted segment of the plot area. Markers entered during acquisition appear just above the plot area; small 'n' buttons along the top or bottom of the plot area (not in this example) indicate notes entered while gathering data; clicking one of these buttons brings up the note text (example):
The major components are the plot area (top half of the screen), yellow
data bar (upper left), screen selection slider (bottom edge of plot area), 'toolbar'
menu buttons (across the center, just below the screen selection
slider -- optionally, the toolbar can be above or to the left of the plot area), comments window (middle right), block window (lower
right), and results window (lower left; this contains controls for EDIT menu operations as well as analysis results). A selected block
appears as a color-inverted segment of the plot area. Markers entered during acquisition appear just above the plot area; small 'n' buttons along the top or bottom of the plot area (not in this example) indicate notes entered while gathering data; clicking one of these buttons brings up the note text (example):

 This
submenu has two items.
This
submenu has two items.